Running asterism selections
📑 Pipeline
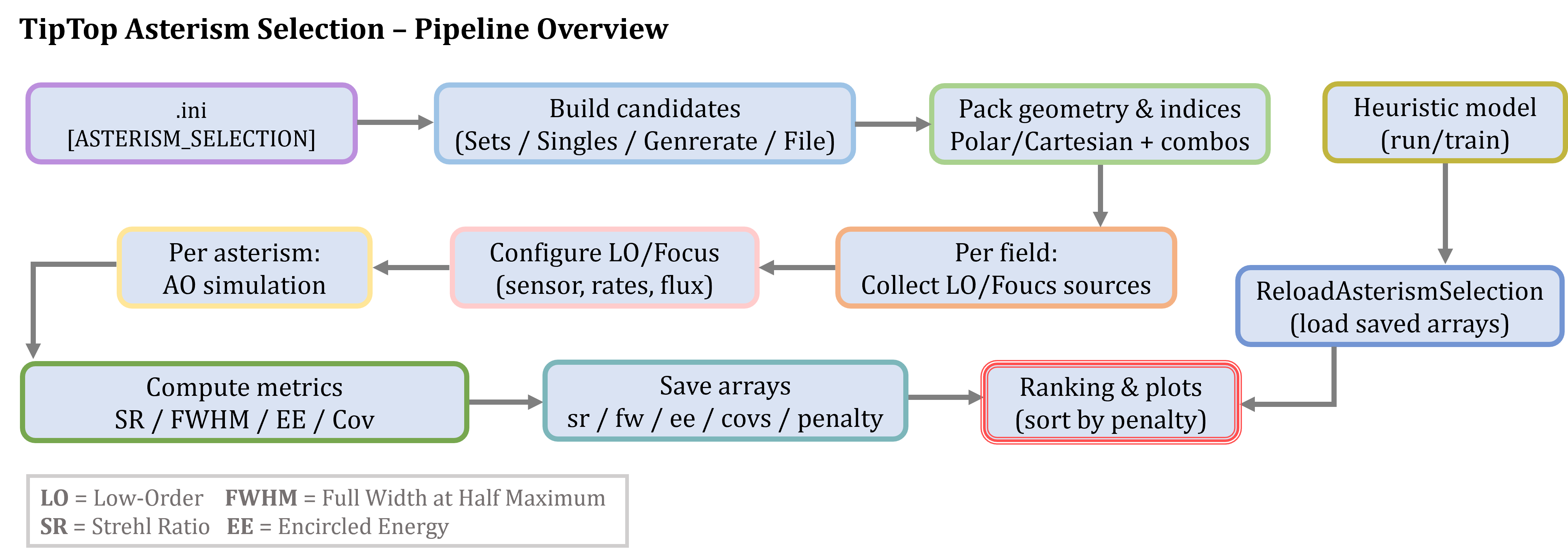
📈 asterismSelection function
Once your .ini has an [ASTERISM_SELECTION] section, run the evaluation with:
tiptop.asterismSelection(simulName, path2param, parametersFile, outputDir, outputFile, doPlot=False,
returnRes=False, returnMetrics=True, addSrAndFwhm=True, verbose=False,
getHoErrorBreakDown=False, ensquaredEnergy=False, eeRadiusInMas=50,
doConvolve=False, plotInComputeAsterisms=False, progressStatus=False, gpuIndex=0):
Parameters (highlights)
Parameters
| Parameter | Type | Required | Description |
|---|---|---|---|
simulName | str | ✓ | Short name used as prefix when saving arrays (e.g. simulName+'sr.npy'). |
path2param | str | ✓ | Path to the parameter file. |
parametersFile | str | ✓ | Name of the parameter file to be used without the extention. |
outputDir | str | ✓ | Path to the folder in which to write the output. |
outputFile | str | ✓ | Base FITS filename if results are saved as images elsewhere in the pipeline. |
doPlot | bool | optional | Default: False, If you want to see the result in python set this to True. |
returnRes | bool | optional | Default: False, If True, return (HO, LO) residuals per asterism and the simulation object. |
returnMetrics | bool | optional | Default: True, The function will return Strehl Ratio, fwhm, encircled energy within eeRadiusInMas, covariance ellipses and the simulation object, if set to True. |
addSrAndFwhm | bool | optional | Default: True, The function will add in the header of the fits file SR anf FWHM for each PSF. |
verbose | bool | optional | Default: False, If you want all messages set this to True. |
getHoErrorBreakDown | bool | optional | Default: False, If you want HO error breakdown set this to True. |
ensquaredEnergy | float | optional | Default: False, If you want ensquared energy instead of encircled energy set this to True. |
eeRadiusInMas | bool | optional | Default: 50, Used together with returnMetrics, radius used for the computation of the encirlced energy. |
doConvolve | bool | optional | Default: False, If you want to use the natural convolution operation set to True. |
plotInComputeAsterisms | bool | optional | Default: False, If you want to display asterisms. |
progressStatus | bool | optional | Default: False, If you want to display progress status. |
gpuIndex | integer | optional | Default: 0, Target GPU index where the simulation will be run. |
⚠️ Prerequisites: your .ini must have a valid [ASTERISM_SELECTION] and the LO loop must be on (valid [sensor_LO] section).
✨ What does asterismSelection produce?
tiptop.asterismSelection(...) evaluates every candidate asterism and returns/saves image-quality metrics per asterism, plus a simulation object you can reuse.
Return values
Depending on the flags, you typically get:
sr, fw, ee, covs, simul = tiptop.asterismSelection(...,
returnRes=False, # set True to also get residuals (see below)
returnMetrics=True # default=True
)
sr: Strehl Ratio per asterism (unitless, ∈ [0,1]).fw: FWHM per asterism (in milliarcseconds, mas).ee: Encircled Energy per asterism, measured within eeRadiusInMas (fraction ∈ [0,1]).covs: covariance ellipses per asterism (per science target; used for anisotropy/jitter visualizations).simul: the simulation object (class asterismSimulation) holding inputs, indices, and cached arrays.
If you pass returnRes=True, the function also returns residual terms (HO/LO) per asterism alongside the simulation object. These are the raw residuals produced by the AO pipeline and are useful for diagnostics (e.g., breaking down error sources). Units and exact shapes depend on your AO configuration (number of science targets, WFS setup, etc.).
✏️ Note on shapes. Metrics are computed per science target.
If you configured multiple science directions, each asterism entry contains one value per target (e.g. sr[asterism_idx, target_idx]). The first target ([..., 0]) is often the “on-axis” metric. With a single science target, arrays reduce to length nAsterisms.
Files written to outputDir
For convenience and faster reloads, the following arrays are saved with the prefix simulName:
| File | Contains | Notes |
|---|---|---|
simulName + 'sr.npy' | Strehl Ratio per asterism (per target) | unitless |
simulName + 'fw.npy' | FWHM per asterism (per target) | mas |
simulName + 'ee.npy' | Encircled energy per asterism (per target) | fraction |
simulName + 'covs.npy' | Covariance ellipses per asterism (per target) | used for PSF anisotropy/jitter |
simulName + 'penalty.npy' | Scalar ranking metric per asterism | lower is better |
⬇️ You can reload them without recomputing via:
sr, fw, ee, covs, simul = reloadAsterismSelection(simulName=SIMUL_NAME,path2param=PARAMS_DIR,
parametersFile=INI_BASENAME, outputDir=OUTPUT_DIR,
outputFile='psf', doPlot=False,
returnRes=False, returnMetrics=True,
addSrAndFwhm=True, verbose=False,
getHoErrorBreakDown=False, ensquaredEnergy=False,
eeRadiusInMas=50, gpuIndex=GPU_INDEX)
For a step-by-step run, see Tutorial - Asterism Seclection.