Tutorials Useful & Scripts
This section provides a collection of useful scripts and detailed tutorials designed to help you run, extend, and better understand TipTop simulations for your own applications.
📝 Tutorials
🔵 Understanding Additional Error Terms in TipTop (Jitter, Extra Error and Static aberrations)
This tutorial introduces the optional parameters that can be activated in TipTop
to enrich simulated PSFs with additional, user-defined error contributions.
These parameters allow you to represent effects that are not otherwise considered by TipTop, such as
non common path aberrations (NCPA), manufacturing tolerances, phasing errors in pupil segmented systems, or residual vibrations, ...
These configurable parameters can help you to produce more realistic PSFs, test “what-if” error budgets, or reproduce measured PSFs for specific analyses.
Part of the purpose of this tutorial is therefore to illustrate which regions of the PSF
(core width, halo level, etc.) are most sensitive to each additional error term.
However, several effects can overlap, and the impact of individual parameters is sometimes
difficult to distinguish by eye. As a result, manual tuning often requires iterating and
revisiting parameters more than once. Because some error terms may compensate or mask each other, caution is recommended.
Before introducing new aberrations, users should refer to the page
“What is included in TipTop?” to avoid adding errors that are already taken into account.
Overview of the main parameters
| Parameter | Required? | Type | Description |
|---|---|---|---|
jitter_FWHM | No | float | Default: None, Additional kernel to be convolved with PSF, it could be a scalar (FWHM in mas) for a round kernel or a list of three values [FWHM_mas_max, FWHM_mas_min, angle_rad]. |
extraErrorNm | No | float | Default: 0, nm RMS of the additional wavefront error to be added (an error that is not otherwise considered). |
extraErrorExp | No | float | Default: -2, Exponent of the power of spatial frequencies used to generate the PSD associated with extraErrorNm. |
extraErrorMin | No | float | Default: 0, Minimum spatial frequency [m−1] for which PSD associated with extraErrorNm is > 0. |
extraErrorMax | No | float | Default: 0, Maximum spatial frequency [m−1] for which PSD associated with extraErrorNm is > 0. Note: 0 means maximum frequency is the one present in the spatial frequency array of the PSDs. |
PathStaticOn | No | string | Default: None, Path to a map of static aberrations (nm) in .fits file. If absent or ‘’, not used. |
zCoefStaticOn | No | list | Default: None, Vector with zernike amplitudes of a static aberration. Coefficients are in nm RMS. If absent not used. |
1. Jitter (jitter_FWHM)
The parameter jitter_FWHM defines a Gaussian kernel (FWHM in mas) that is convolved with the final PSF.
It is used to model an additional tip/tilt jitter ( e.g. vibrations, PSF drifts, ...).
✏️ Implementation note:
This Gaussian convolution is applied at the final stage of
TipTop’s pipeline (in tiptop.baseSimulation.finalConvolution()).
How to define it:
- Circular jitter
[telescope]
jitter_FWHM = 30.0 ; in mas
- Elliptical jitter:
[telescope]
jitter_FWHM = [25.0, 20.0, 30.0] ; FWHM_X, FWHM_Y [mas], angle [deg]
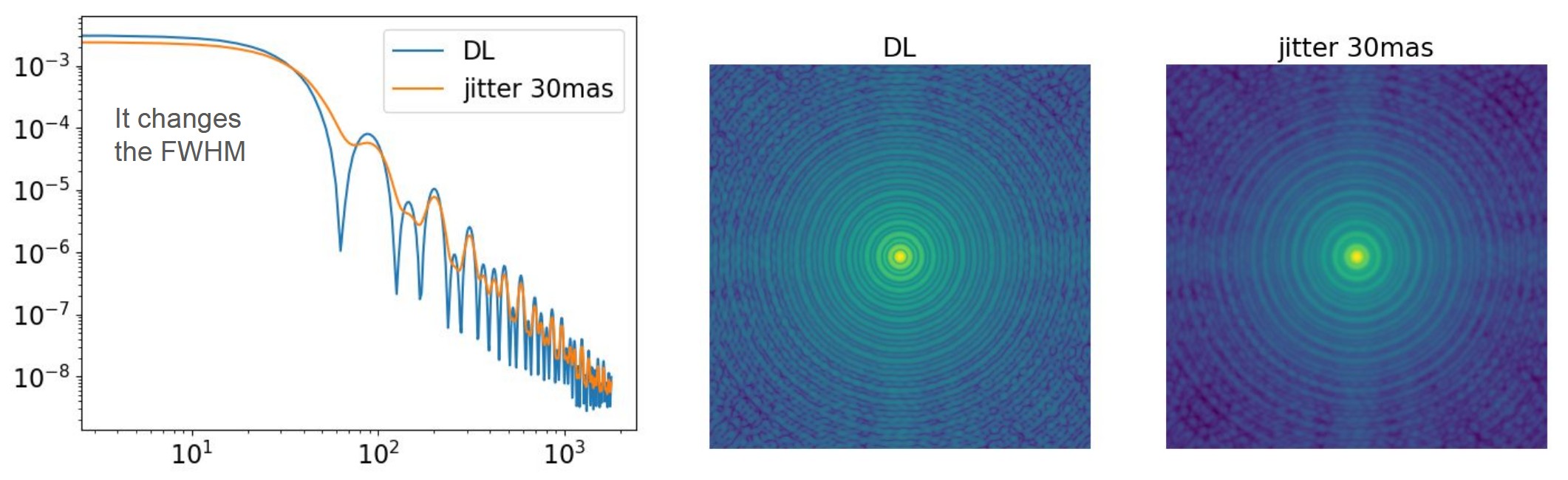
Effects of jitter_FWHM :
- Broadens the PSF core (direct effect).
- Increases the PSF FWHM as the jitter amplitude grows.
- Reduces the Strehl ratio, since energy leaves the core.
- Slightly smooths the halo
Because jitter_FWHM corresponds to tip–tilt–dominated residuals, it mainly acts on
the lowest spatial frequencies: it widens the core while having minimal effect on the
shape of the outer AO halo.
Tuning
In practice, this extra parameter is often added first, then its value is refined after introducing extraError* and
zCoefStaticOn, since these parameters also modify the core–halo energy balance
(although through different mechanisms).
2. Extra high-order error (extraErrorNm, extraErrorExp, extraErrorMin, extraErrorMax)
The extraError* parameters allow you to introduce an additional, user-defined wavefront error
described by a simple spatial-frequency power law.
This term is useful to model high-order aberrations not otherwise represented in TipTop, such as
residual manufacturing erros, residual telescope contributions, or any unmodelled generic high-order
static term.
✏️ Implementation note:
TipTop computes an additional PSD from these
parameters inside p3.aoSystem.fourierModel.extraErrorPSD(), and this PSD is then
added to the AO PSD in p3.aoSystem.fourierModel.powerSpectrumDensity()
(after aliasing, noise, temporal and other contributions).
This provides a straightforward way to inject a generic static high-order
aberration PSD into the error budget.
Meaning of the parameters
extraErrorNm– total RMS amplitude (in nm) of the additional errorextraErrorExp– exponent of the power-law spectrum (default –2)extraErrorMin/extraErrorMax– optional lower/upper spatial-frequency limits (in m−1).
Effects of the extraError* parameters
- Strehl ratio reduction
IncreasingextraErrorNmincreases the total WFE and transfers energy from the PSF core into the halo. - Encircled-energy modification
HigherextraErrorNmvalues decrease EE at small radii and increase it at larger radii. - Halo-shape variation
The power-law exponentextraErrorExpcontrols how energy is distributed across spatial frequencies:- steeper (more negative) slopes → more low-frequency power → broader halo
- shallower slopes → more high-frequency power → sharper halo edge
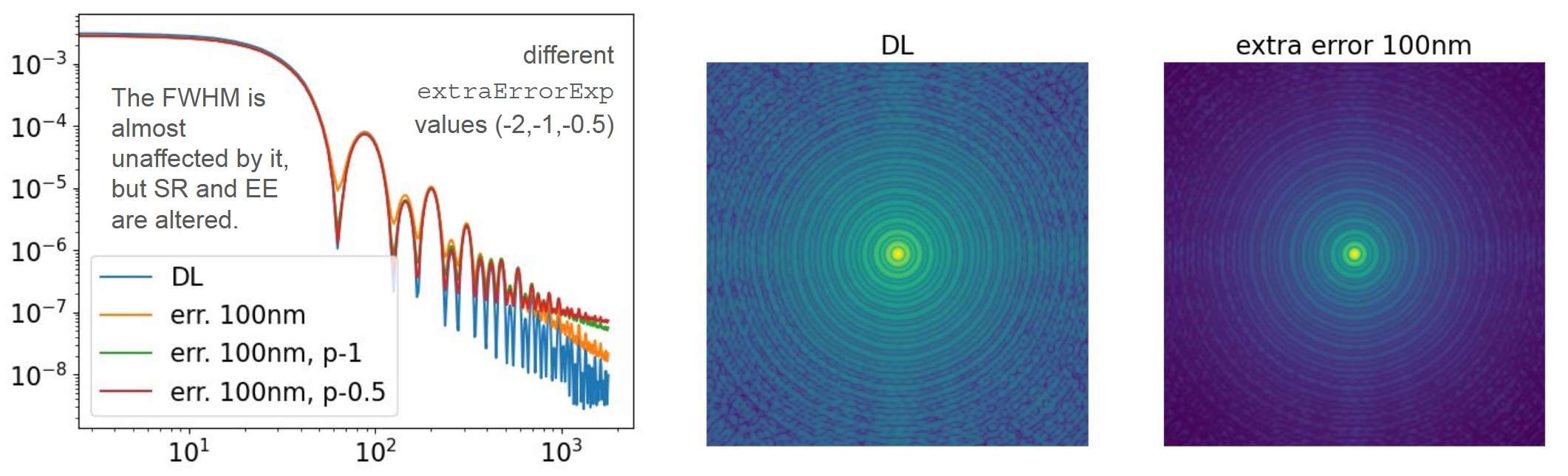
The exponent extraErrorExp determines how quickly the PSD decreases with frequency.
Changing this exponent modifies how energy is redistributed between intermediate and high spatial frequencies, which directly affects the core–to–halo balance.
extraErrorMin and extraErrorMax further allow you to restrict
the spatial-frequency range over which the additional error is applied.
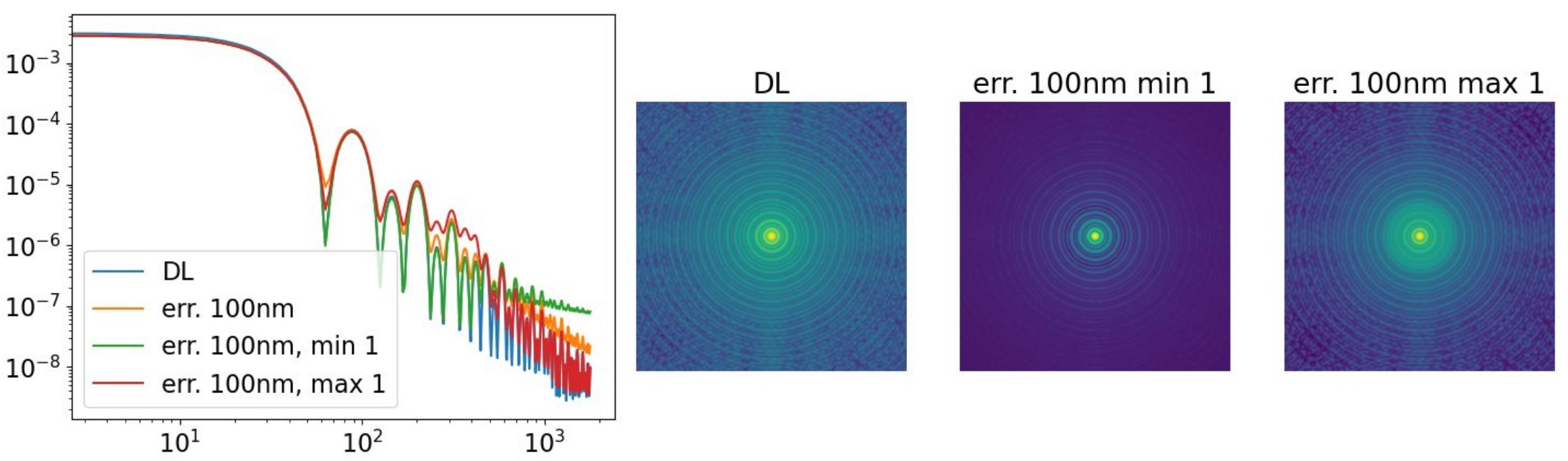
Tuning
In practice, after adding jitter_FWHM, the
extraError* parameters are typically used to adjust the level and shape of the
halo.
A larger extraErrorNm decreases the Strehl ratio by injecting
additional high-order wavefront error, thereby raising the halo.
The exponent extraErrorExp then provides finer control over the
halo slope by redistributing energy between intermediate and high spatial
frequencies.
Because the effects of several parameters can partially overlap, the tuning process is
iterative: modifying extraErrorNm often requires
revisiting extraErrorExp, and these adjustments must remain
consistent with any static aberrations introduced through
zCoefStaticOn or PathStaticOn.
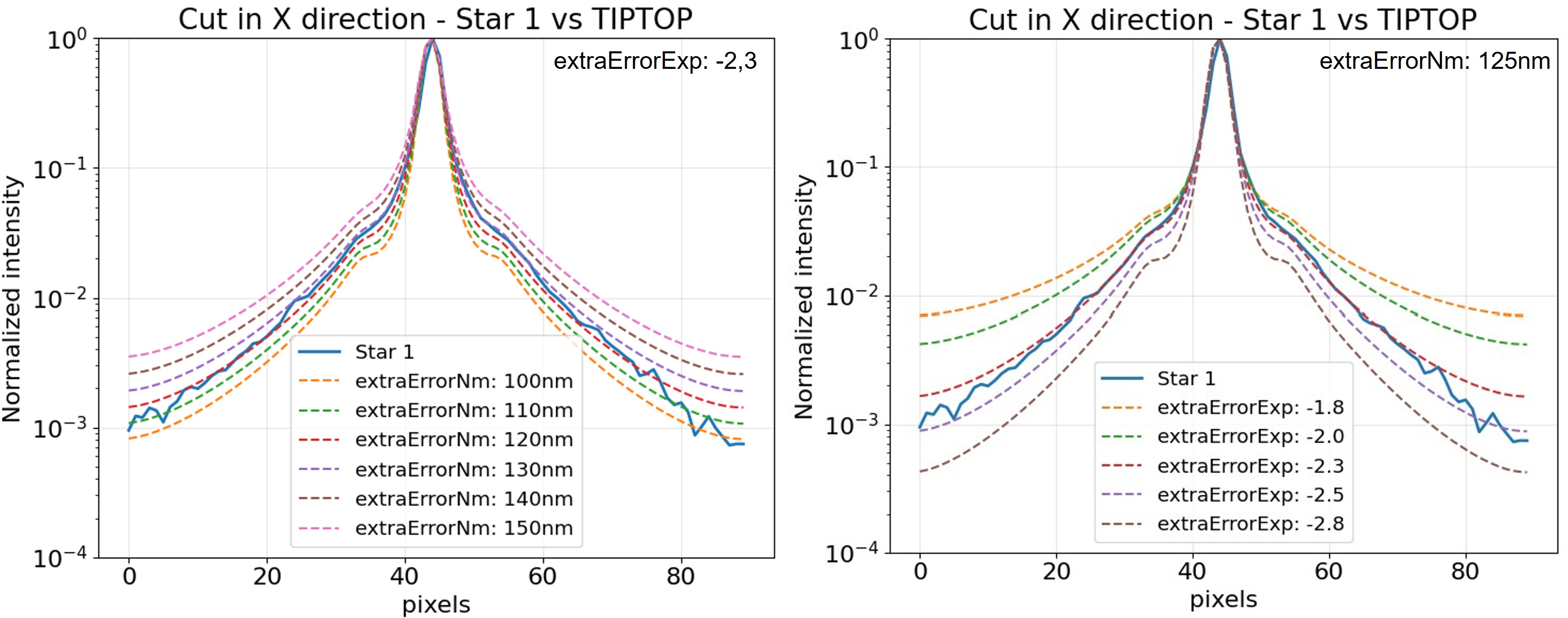
In the "MUSE PSF-fitting" example shown below (for a science wavelength of 500nm),
the effect of extraErrorNm and extraErrorExp
on the radial and directional profiles can be inspected to determine which combination best reproduces the observed halo amplitude and slope.
The plots illustrate how varying these parameters affect the PSF shape during manual tuning.
For simplicity, only the X-cut of the PSF is shown here for a single star —
the objective is purely to visualize the qualitative impact of these parameters.
Note: the example results correspond to simulations using zCoefStaticOn = [0, 0, 100] (focus term added).
3. Static aberrations (PathStaticOn, zCoefStaticOn)
Static aberrations correspond to wavefront errors that are fixed in time and do not evolve
during the simulation.
In TipTop, to introduce such static terms you can define:
- a user-provided OPD map (
PathStaticOn) - a set of Zernike coefficients (
zCoefStaticOn)
✏️ Implementation note:
Both contributions are combined into an on-axis OPD map (in nm) inside the
telescope class (attribute tel.opdMap_on).
This OPD map is then applied in the pupil plane when computing the final PSFs, on top of the AO residual
wavefront error.
3.1 Using an OPD map (PathStaticOn)
PathStaticOn allows you to load a static optical path difference map (in nm)
from a FITS file.
It adds a static aberration (it can be used to add any kind of static aberrations).
This is the recommended approach when you have a measured or simulated aberration map.
The OPD map is rescaled/rotated to match the telescope pupil resolution and geometry and is then
used as an additional OPD term during PSF computation.
One example is the static aberration given by ELT M1 (available here).
![]()
3.2 Using Zernike coefficienrs (zCoefStaticOn)
zCoefStaticOn provides a compact way to introduce a combination of zernike modes that model a static aberration.
The values are given in nm RMS and follow the standard Noll index ordering (Z2 = tilt, Z3 = tilt, Z4 = defocus, Z5–Z6 = astigmatism, …).
✏️ Implementation note:
Inside telescope class, TipTop:
- builds the corresponding Zernike basis on the pupil,
- orthonormalizes the modes on the actual pupil support,
- normalizes each mode to 1 nm RMS,
- and finally forms
opdMap_onby summing the modes weighted by the coefficients inzCoefStaticOn.
Examples:
| Mode | zCoefStaticOn |
|---|---|
| Focus | [0, 0, 100] |
| Astigmastism | [0, 0, 0, 100] |
| Trefoil | [0, 0, 0, 0, 0, 0, 0, 100] |
| Spherical | [0, 0, 0, 0, 0, 0, 0, 0, 0, 100] |
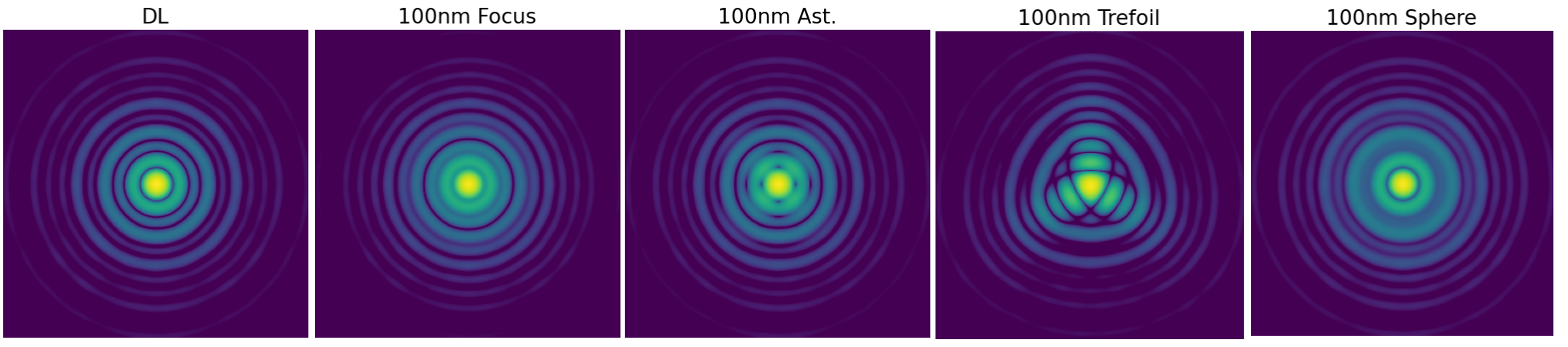
Effects of static aberrations
- Strehl ratio degradation — depending on the total RMS and the Zernike content.
- PSF shape modification — Zernike modes introduce directional features (e.g. astigmatism, coma), while arbitrary OPD maps can create more complex patterns.
- Energy redistribution — static aberrations alter both the PSF core and halo, with characteristic signatures depending on the aberration type.
Typical use
Use PathStaticOn when you have a known, structured pupil-plane aberration
(e.g. calibration-derived NCPA, optics maps, segmented mirror errors).
Use zCoefStaticOn to explore the impact of specific modes (focus, astigmatism, trefoil, …)
or to test simple static error scenarios in a controlled way.
Tuning
In our "MUSE tuning example", a set of Zernike coefficients was used together with jitter_FWHM
and extraError* to reproduce the observed PSF, matching both core width and halo level.
The example below illustrates the impact of adding a simple defocus term via
zCoefStaticOn = [0, 0, 100] on the PSF shape (X-cut only, for clarity).
![]()
Note on wavelength: the sensitivity of PSF features to these parameters depends on the simulation wavelength. Users should perform comparisons at the wavelength(s) most relevant for their science case, as different wavelengths emphasise different spatial frequency ranges of the PSF.
📄 Useful Scripts
🔵 Run N TipTop Simulations with Different Parameter Values
Here, we provide an example script to run N TipTop simulations with different values of two parameters:
ZenithAngle and Seeing. The script can easily be adapted to sweep over a single parameter or more than two.
Download the script: 📥.
Running N TipTop simulations: TIPTOP_RUN_N_SIM.py
"""
Created on Thu Jul 10 10:00:34 2025
Run N TIPTOP simulations by sweeping over different parameter values.
@author: astro-tiptop-services
"""
#%% =============================================================================
# Import necessary libraries
from tiptop.tiptop import overallSimulation
import numpy as np
import configparser
from itertools import product
import os
#%% =============================================================================
# === File Paths ===
#💡 Adapt these paths to your environment
inputDir = "../MICADO/" # Folder containing the .ini parameter file
inTag = "MICADO_SCAO" # Name of the input parameter file (without extension)
outputDir = "./" # Folder where the output FITS files will be saved
tempDir = inputDir # Temporary folder for modified .ini files
tempName = 'file_temp' # Name of the temporary file
# Construct the full path to the input and temporary config files
inputFile = os.path.join(inputDir, f"{inTag}.ini")
tempFile = os.path.join(tempDir, f"{tempName}.ini")
# Base name for output files
outTagBase = "MICADO"
#%% =============================================================================
# === Define parameters to sweep ===
# 💡 Format: (section_name, key_name, list_of_values : min, max, number of values)
param_sweep = [
("atmosphere", "Seeing", np.linspace(0.8, 2.0, 3)),
("telescope", "ZenithAngle", np.linspace(0.0, 60.0, 3)),
# Add more parameters as needed
]
#%% =============================================================================
# === Read base configuration file ===
parser = configparser.ConfigParser()
parser.optionxform = str # Keep the original case of parameter keys
parser.read(inputFile)
#%% =============================================================================
# === Build and run combinations ===
# Extract parameter names and values
param_names = [f"{sec}.{key}" for sec, key, _ in param_sweep]
param_values = [vals for _, _, vals in param_sweep]
# Generate all combinations
for combo in product(*param_values):
# Update .ini with current parameter values
for (section, key, _), value in zip(param_sweep, combo):
parser.set(section, key, str(value))
# Write updated config to temporary .ini file
with open(tempFile, "w") as configfile:
parser.write(configfile)
# Build output file tag
tag_parts = [f"{name.split('.')[-1]}{val:.2f}" for name, val in zip(param_names, combo)]
tag = f"{outTagBase}_{'_'.join(tag_parts)}"
print(f"🔄 Running simulation with: {dict(zip(param_names, combo))}")
# Run TipTop simulation
overallSimulation(
path2param=tempDir,
parametersFile=tempName,
outputDir=outputDir,
outputFile=tag,
addSrAndFwhm=True
)
🔵 Rename Output FITS Files and Keep Backups
This script renames PSF FITS files (e.g., those generated by the previous script)
to facilitate their use in other routines, while creating backups of the originals.
For example, it renames FITS files in your folder starting with the tag MICADO
to simple, easily readable names like MICADO_PSF1.fits, MICADO_PSF2.fits, and so on.
Download the script: 📥
Rename output PSF FITS files: TIPTOP_rename_psf_files.py
"""
Created on Thu Jul 10 14:15:45 2025
Rename output PSF FITS files sequentially for easier access and keep original copies.
@author: astro-tiptop-services
"""
#%% =============================================================================
# Import necessary libraries
import os
import shutil
#%% =============================================================================
# === Configuration ===
#💡 Adapt these paths to your environment
inputDir = "./" # Directory containing the generated FITS files
backupDir = "Original_PSFs" # Directory to store backups of original files
outputDir = "Renamed_PSFs" # Directory where renamed files will be moved (must exist)
TagBase = "MICADO" # Base name used in the output FITS filenames
# Create output and backup directories if they do not exist
os.makedirs(outputDir, exist_ok=True)
os.makedirs(backupDir, exist_ok=True)
#%% =============================================================================
# === Gather and sort relevant FITS files ===
output_files = sorted(
[f for f in os.listdir(inputDir) if f.startswith(TagBase) and f.endswith(".fits")]
)
# Check if any matching files are found
if not output_files:
print("[INFO] No matching FITS files found for renaming.")
else:
print(f"[INFO] Found {len(output_files)} FITS files to rename.")
#%% =============================================================================
# === Copy and Rename files sequentially as TagBase_PSF1.fits, MICADO_PSF2.fits, etc. ===
for i, fname in enumerate(output_files):
src = os.path.join(inputDir, fname)
dst = os.path.join(outputDir, f"{TagBase}_PSF{i+1}.fits")
backup = os.path.join(backupDir, fname)
try:
# Copy the original file to the backup folder
shutil.copy2(src, backup)
# Rename (and move) the file to the new destination
os.rename(src, dst)
print(f"[INFO] Renamed: {fname} → {TagBase}_PSF{i+1}.fits (Backup saved)")
except PermissionError:
print(f"[⚠️ WARNING] File in use and cannot be renamed: {fname}")
except Exception as e:
print(f"[❌ ERROR] Failed to rename {fname} → {e}")
#%% =============================================================================
print(f"\n✅ Process complete. {len(output_files)} files renamed and backed up.")
🔵 Sum rotated PSF
This script combines multiple PSFs into a single image by rotating each individual PSF
by a specified field rotation angle before summation.
Used together with the two previous scripts, this process can be
especially useful, for example, when simulating long integrations to reproduce the impact of pupil rotation and field rotation on the final image.
✏️ Note: You need to manually define the list of field rotation angles in the script — one angle per PSF FITS file.
Feel free to adapt this script to suit TipTop simulations involving multiple science sources.
The example provided here illustrates its use with the input parameter file
MICADO.ini.
Download the script: 📥
Sum rotated AO PSFs: TIPTOP_rotated_PSF_sum.py
"""
Created on Thu Jul 10 15:01:55 2025
Sum rotated PSFs.
Each PSF is rotated by a specified angle before summation.
@author: astro-tiptop-services
"""
#%% =============================================================================
# Import necessary libraries
import os
import numpy as np
from scipy.ndimage import rotate
from astropy.io import fits
#%% =============================================================================
# === Configuration ===
inputDir = "Renamed_PSFs" # Folder where the PSF FITS files are stored
TagBase = "MICADO" # Base tag for the PSF files
outputFile = "PSF_sum.fits" # Filename for the combined output
# Rotation angles (degrees) applied to each PSF before summation
rotation_angles = [...] # 💡 To be defined
N = len(rotation_angles) # Should match the number of PSF files available
#%% =============================================================================
# === Load and combine rotated PSFs ===
first_filename = os.path.join(inputDir, f"{TagBase}_PSF1.fits")
if not os.path.exists(first_filename):
raise FileNotFoundError(f"Base file {first_filename} is missing. Check input directory.")
first_img = fits.getdata(first_filename)
accumulated = np.zeros_like(first_img, dtype=float) # Accumulator for summing rotated PSFs
valid_count = 0 # Count how many files were found and used
# Loop over all images and angles
for i in range(N):
filename = os.path.join(inputDir, f'{TagBase}_PSF{i+1}.fits')
# Check if file exists
if not os.path.exists(filename):
print(f"[WARNING] Missing file: {filename}")
continue
print(f"[INFO] Loading file: {filename}")
img = fits.getdata(filename)
# Rotate the image by the corresponding angle
rotated = rotate(img, angle=rotation_angles[i], reshape=False)
# Accumulate the rotated image
accumulated += rotated
valid_count += 1
#%% =============================================================================
# === Normalize and save result ===
if valid_count > 0:
accumulated /= valid_count
# Save PSF sum to a FITS file, overwrite if exists
fits.writeto(outputFile, accumulated, overwrite=True)
print(f"[INFO] Combined PSF saved to: {outputFile}")
else:
print("[ERROR] No valid PSF files found. No output generated.")